Statistics and the Origin of Life
A 2009 New Scientist article stated:
There is no doubt that the common ancestor (the first life form) possessed DNA, RNA and proteins, a universal genetic code, ribosomes (the protein-building factories), ATP and a proton-powered enzyme for making ATP. The detailed mechanisms for reading off DNA and converting genes into proteins were also in place. In short, then, the last common ancestor of all life looks pretty much like a modern cell.
The term ‘common ancestor’ is an abbreviated form of biochemical term ‘The Last Universal Common Ancestor (LUCA)’. If we were able to trace our ancestry back to the first cell, LUCA is the cell that would be the end of our ancestry line, i.e. the stem of our ancestry tree.
LUCA is the minimum requirement for life.
The New Scientist, a secular magazine, would have had no incentive to describe the earliest life form as ‘complex’. Evolution theory requires the first cell to be simple.
If random chance is the origin of life, this first living cell logically needs to be primitive, or in other words, an easily explained, simple cell. However, after the New Scientist embarked on an investigation to define the simplest cell possible and the minimum requirement for life, the evidence compelled them to describe their findings scientifically.
The detail
LUCA turns out to be a cell with stunning detail, and which also contains a vast amount of sub-components and ‘machinery’, in addition to some very complex chemistry. A sample of this exquisite detail may be viewed on the videos below
Electron transport: https://m.youtube.com/watch?v=rdF3mnyS1p0&feature=youtu.be
How enzymes work: https://m.youtube.com/watch?feature=youtu.be&v=yk14dOOvwMk
From DNA to protein: https://m.youtube.com/watch?v=D3fOXt4MrOM&feature=youtu.be
Silencing rna: https://m.youtube.com/watch?v=t5jroSCBBwk
The New Scientist found no indication of a ‘primitive cell’, nor of any simplicity at all. Neither did they find a discernible difference from the ‘modern cell’.
The question now is: ‘What are the chances of such an ancestral cell coming into existing by purely random processes?’ If God did not create, but random chance is the origin of life, what are the statistics for such an accidental occurrence? What is the probability for the first and most simple cell to have formed spontaneously?
The bacterium Mycoplasma genitalium has the smallest known genome of any known organism . Mycoplasma contains 482 genes comprising 580,000 bases. Of course, these genes are only functional with pre-existing translational and replicating machinery, a cell membrane, etc. But Mycoplasma genitalium has no cell walls, and can only survive by parasitizing more complex organisms (e.g. it lives in the cells of the respiratory system and urogenital tract of humans) that provide many of the nutrients it cannot manufacture for itself. Indeed, this organism seems to have arisen by loss of genetic information, making it dependent on a host. The mycoplasmas are very simple bacteria (although still quite complex). Could anything ‘simpler’ have arisen?
The bare minimum
A decade ago, Eugene Koonin, a researcher interested in making artificial biological organisms, tried to calculate the bare minimum required for a living cell. He based his work on the mycoplasmas, and estimated how many genes even these simple cells could do without. His team came up with a result of 256 genes. They doubted whether such a hypothetical bug could survive for long, because such an organism could barely repair DNA damage, could no longer fine-tune the ability of its remaining genes, would lack the ability to digest complex compounds, and would need a comprehensive supply of organic nutrients in its environment. It is not surprising that follow-up research has revised this number significantly upwards. This new hypothetical minimum genome consists of 387 protein-coding and 43 RNA-coding genes.
LUCA
In 2011, evolutionary biologists discussing this hypothetical Last Universal Common Ancestor (LUCA) likewise realized such a thing could not have been simple. Rather, it would have included a ‘universal organelle’ to store high energy compounds called pyrophosphates; previous dogma held that bacteria lacked organelles. A report stated: New evidence suggests that LUCA was a sophisticated organism after all, with a complex structure recognizable as a cell, researchers report. No origin-of-life simulation approaches this bare minimum in the slightest.
Could this complexity have arisen by chance? Natural selection can’t operate without a self-sustaining, reproducing system. Therefore, natural selection can’t be invoked to explain this minimum level of complexity. All evolutionists have is chance, which is amenable to fairly simple probability calculations. Information theorist Hubert Yockey calculated that, given a pool of pure, activated biological amino acids (a much more generous offer than the hypothetical ‘primordial soup’), the total amount of information which could be produced, even allowing for a billion years of trial and error, as evolutionists posit, would be only a single small polypeptide, 49 amino acid residues long. This is about 1/ 8 the size (therefore 1/8 the information content) of a typical protein.
1/8th (0.125) of a protein
A billion years of trial and error could statistically produce about 1/ 8 of a typical protein, yet the hypothetical simple cell LUCA needs at least 387 proteins, all pre-coded in DNA, and that would only allow it to live in a very specific and invariant environment with a constant supply of high-level nutrients and biomolecules. And Yockey’s estimate generously presupposes that the many chemical hurdles can be overcome, which is a huge assumption, as will be shown later.
Alternatively, one could calculate the probability of obtaining the DNA sequences for each of these proteins at random. Certainly there is some leeway in the sequence for many, but not around the active sites. However, even evolutionary writers implicitly concede that some sequences are essential. They call them ‘conserved’.
Fortunately, calculation of random chance occurrences is a well developed branch of mathematics, called statistics. Using statistics, given a certain set of constituent components and by a simple addition process, the probability of the spontaneous occurrence of a certain combination of cell components can be calculated.
The statistics.
Anyone who has rolled a dice will know that the probability of throwing a six is 1:6. The statistic is one in six. Similarly, with cards, drawing or dealing the ace of spades from a pack of 52 cards is 1:52, and your chances being dealt any ace is 1:13. When playing poker your chances of a royal flush (ACE, King, Queen, Jack and 10 of hearts) is 52+51+50+49+48 = 1:250
The purpose of the math below is to show how the scientific advances in the understanding of life processes on cellular level have stunned the scientific world.
The chemistry of the simplest cell is unimaginably complex. Random chance seems insufficient to produce the basic minimum requirements necessary for life. As we will calculate below, there are 10^5034 random interactions needed for the random formation of the chemistry of the simplest life form. Comparing this to the calculated statistic of 10^110 atomic interactions that have been possible since the origin of the universe (see below), it seems that the theorized billions of years since the big bang are hopelessly inadequate to explain the statistics required for the random formation of the simplest life form.
The simplest life form would consist of: (explained in more detail later in this post)
- An abundant supply of the 20 different amino acids, constituting the protein based ‘building blocks’ of life.
- 387 proteins for the simplest possible life with 10 conserved amino acids on average. (definition of ‘conserved’ to follow)
∴ The chance for the random formation of the required 387 proteins is 20^–3870 = 10^–(3870. log20) = 10^–5035
This is one chance in one followed by over 5,000 zeroes, i.e. one number 5000 digits in length. In lay terms, it would be like guessing a PIN number of 5000 digits at the first go. This one-off code number would be so long that it would take 20 pages of 250 numerals per page to write it down, and with only one chance of guessing it correctly.
The theorized billions of years since the big bang is simply a too infinitesimally small a time for random chance to create the chemistry of the simplest life form. To prove that consider the following statistics:
- There are 10^80 atoms in the universe.
- There are 10^18 seconds elapsed since the origin of the universe. (according to the big bang theory)
- There are 10^12 atomic interactions possible per second per atom
Doing the maths (10^(80+18+12)) shows that ∴ only 10^110 atomic interactions have been possible since the origin of the universe.
As we start suspecting, science, chemistry and statistics have not supported, but have rather played the role to rule out random chance as an ‘origin of life’ theory. If that is not enough, a further component not yet considered- information-, is the main problem for the ‘random chance’ origin of life theory. In addition to the random occurrence of the right chemistry, as described above, information transfer and maintenance pose further challenges, which are even more daunting. Even the simplest imagined life would require an enormous information content. (a virus doesn’t count because it is utterly dependent on the machinery of more complex cells for reproduction and assembly). View the Scientific American video https://www.youtube.com/watch?v=C8OL1MTbGpU for some basic information on DNA, genes and chromosomes.
Conserved
Conserved (essential)whole proteins include the histones that act as spools around which DNA wraps in chromosomes. In addition, conserved proteins also include ubiquitin, which is ubiquitous in organisms apart from bacteria and essential for marking unwanted proteins for destruction, and calmodulin, the ubiquitous calcium-binding protein which has almost all of its 140–150 amino acids ‘conserved’.
Discussion on the Math
The following calculation will be very generous to evolutionists. We will pretend that only 10 amino acids are conserved per enzyme and that there is some mechanism for joining amino acids in solution to form long chains (this is overly generous, as water constantly hydrolyses peptide bonds in solution): 20 amino acids 387 proteins for the simplest possible life 10 conserved amino acids on average ∴ chance is 20^–3870 = 10^–3870. log20 = 10^–5035 This is one chance in one followed by over 5,000 zeroes. So it would be harder than guessing a correct 5,000-digit PIN on the first go! Yet, without this entirely fortuitous assemblage, life is not possible. This is not a matter of slowly building up pre-living chemicals, for this is the simplest form of life according to those who believe the origin of life is possible from a chemical soup. Many evolutionists have said that, given enough time, anything is possible. However, is time really ‘the hero of the plot’? No. There are:
10^80 atoms in the universe,10^12 atomic interactions per second, 10^18 seconds since the origin of the universe, according to the big bang theory ∴ only 10^110 interactions are possible.
This is a huge number, but compared with the number of trials necessary to have a reasonable chance of obtaining the right sequence of nucleotides required to code for the necessary proteins required by the simplest conceivable life form, it is absurdly small. Even given these reactions, there would be but one chance in 10^4925. These numbers are so large as to be meaningless. The point is to illustrate the statistical impossibility (beyond ‘improbability’) of the origin of life from non-living chemicals.
Sir Fred Hoyle
The famous cosmogonist, Sir Fred Hoyle (1915–2001), abandoned his atheism when he considered the absurdly small probabilities: The likelihood of the formation of life from inanimate matter is one to a number with 40,000 naughts after it … It is big enough to bury Darwin and the whole theory of evolution. There was no primeval soup, neither on this planet nor any other, and if the beginnings of life were not random, they must therefore have been the product of purposeful intelligence. Self-replicating molecules?
To try to grasp the above analysis on the minimal complexity of life, some evolutionists have theorized that one type of molecule could perform both catalytic and reproductive roles. The choice is usually between the nucleic acids (RNA) and proteins. However, even now, evolutionists have to admit that RNA is really a lousy catalyst, and proteins are lousy replicators. No RNA enzyme has approached anything like the efficiency of the proteinaceous enzymes needed for living creatures, such as those Dr Wolfenden analyzed, and life as we know it is impossible without those efficiencies. It is no use theorizing about alternate possible life forms either, for we have to explain life as we see it here on planet earth, and that life defies naturalistic stories of origins.
Proteins hopeless at replication
Evolutionists admit that ribozymes (RNA enzymes manufactured in laboratory experiments) are not efficient enzymes; they could never achieve the efficiencies of the enzymes necessary for life. Similarly, even Dawkins had to admit: Darwin, in his ‘warm little pond’ paragraph, speculated that the key event in the origin of life might have been the spontaneous arising of a protein, but this turns out to be less promising than most of Darwin’s ideas. … But there is something that proteins are outstandingly bad at, and this Darwin overlooked. They are completely hopeless at replication. They can’t make copies of themselves.
This means that the key step in the origin of life cannot have been the spontaneous arising of one protein molecule.”
Summary
For those who have eyes to see, it becomes clear that science and chemistry have been major tools operating in favour of ‘intelligent design’. Science has assisted and validated the creation model rather than disprove it.
However, attempting to convince the blind is futile. Any internet search on LUCA will confirm this. Although admittance to the ‘minimum requirement’ is universal, the statistical impossibilities thereof are ignored.
Instead vague unsubstantiated claims about thermal gradients and alternative energy sources in an oxygen free environment are discussed. Insignificant molecule finds at deep-sea hydro-thermal vents dominate the ‘scientific’ reports. None of these molecules even slightly resemble one of the approximately 387 ‘minimum proteins’ required for life. Firstly, for example, there is the admittance to the ‘minimum requirement, as phys.org (2018 article) admits:
Once they had finished their analysis, Bill Martin’s team was left with just 355 genes from the original 11,000, and they argue that these 355 definitely belonged to LUCA and can tell us something about how LUCA lived. (as per https://phys.org/news/2018-12-luca-universal-common-ancestor.html🙂
Then follows an embarrassing flow of vague and unsubstantiated hypothetical illustrations with absolutely nothing but unrealistic postulations. The NY times reports, after an interview with Dr. Sutherland, who chided and exposed the superficiality of Dr Martin deep sea scenarios:
“But chemists like Dr. Sutherland say they are uneasy about getting prebiotic chemistry to work in an ocean, which powerfully dilutes chemical components before they can assemble into the complex molecules of life. …“We didn’t set out with a preferred scenario; we deduced the scenario from the chemistry,” he said, chiding Dr. Martin for not having done any chemical simulations to support the deep sea vent scenario. Dr. Martin’s portrait of Luca “is all very interesting, but it has nothing to do with the actual origin of life,” Dr. Sutherland said.” https://www.nytimes.com/2016/07/26/science/last-universal-ancestor.html
They are truly hoping against hope. Their battle is lost, because the further science advances, the more impossibilities are discovered. It seems that, whether we study microscopically or macroscopically (i.e. the cosmos), that the amount of variables only increases the deeper we dig. The more we find out, the less we know. The more science finds out, the less evolution is supported.
Yet the deception of evolution is promoted at academic and scientific institutions, misleading the man on the street. It is only the light of the gospel that can break the darkness. The two-edged sword of Jesus’s words cut through the lies and deception. As Jesus says: “The words that I speak to you, they are spirit, and they are life.” John 6:63. John says about Jesus earlier on: “In him was life; and the life was the light of men. And the light shines in darkness; and the darkness comprehended it not.” (John 1:4-5)
Only once we place our faith in the God of the bible and in Jesus’s words of life (refer to appendix F), can we truly begin to comprehend the ‘big picture’ and the wonder of life.
*Excerpt from article, “The Origin of Life” by Dr Jonathan Sarfati, PhD Physical Chemistry, in the book “Evolution’s Achilles heels”. References as listed by Sarfati in the book “Evolution’s Achilles heels”.
Disturbing Developments: Gene editing.
Once one has grasped the magnificence and perfection of God’s creative work contained in the human cell, it seems incomprehensible that anyone would dare interfere with it. Every cell contains God’s blueprint for life. Every instruction for the construction of every muscle, nerve, vessel, organ and bone is contained in one cell. The accuracy with which these coded instructions are carried out is incomparable.
For mankind to give itself the right to now manipulate this instruction set designed by God seems truly arrogant in the extreme. Have we got any idea what the consequences are going to be?
Yes, some DNA damage can occur from radiation which does cause mutations, and it seems noble to try and correct these. However, we are moving into dangerous territory, and considering mankind’s history, it seems that evil men will end up exploiting the intentions of the good. From that perspective there is no doubt the commercial considerations will push the limits of ethics and will eventually transgress into human genetic engineering.
Damage repair.
It is furthermore interesting to note that, even though dangerous radiation damages our DNA strands on a daily basis, there is a backup system in place to repair it. Our bodies, and the DNA blueprint thereof, are in a continuous state of monitoring by chemical ‘machines’, which then repair damage or irregularities with the backup system as reference.
How anyone can imagine that a self repair system and a layered backup system of such complexity could develop randomly is inconceivable, yet that is what secular scientists seem to believe.
________________________________________________________
Further Reading for those interested: ( Regarding ‘Junk DNA’, 4-D genetics, splicing, etc)
Firstly some further videos on the amazing cell and DNA detail:
https://www.youtube.com/watch?v=FJ4N0iSeR8U
https://www.youtube.com/playlist?list=PL1ZMVRPLCECalIakXxsePSlf6jcnBrvY7
https://www.youtube.com/playlist?list=PLzKWCpOrkt4QXSOXn3nGk5tbR9wSL9LBk
Junk DNA:
An internet search on the 2003 human genome project will yield the term ‘Junk DNA’. Initially, because the genome researchers could only find 20 000 coded genes, comprising only ~2% of the total amount of genetic material, the remaining ~98% seemed to be unused DNA. This 98% was termed ‘Junk DNA’, as it had no apparent purpose. The irresponsible (and probably mildly arrogant) use of the term ‘junk’ is coming back to haunt those initial researchers. Even though this term is now discarded by scientists, it lives on in the thinking of many laymen. Sarfati continues:
“We have heard this reasoning for years. It goes something like, “Only 2 to 3 percent of the human genome is functional. The rest is worthless, junk DNA—garbage left over from our evolutionary heritage.” Although this is still a commonly-held belief, recent discoveries have shown it to be wrong.
Why, then, do we continue to hear about it so often and for so long?
Is it because biological evolution needs ‘junk DNA’ to solve a great mathematical problem? The problem was already identified by J.B.S. Haldane in the 50’s.
Haldane’s Dilemma
In the late 1950s, the famous population geneticist J.B.S. Haldane showed that natural selection cannot possibly select for millions of beneficial mutations, even over the course of human evolutionary history. Based on observed mutation rates, our common chimp ancestor could only have undergone a few hundred beneficial mutations. Despite employing several simplifying assumptions in favor of evolutionary theory, only a few hundred beneficial mutations could have been available for selection by the ‘natural selection process’ since our common chimp ancestor.
This has become known as Haldane’s Dilemma and, despite many claims to the contrary, Haldane’s Dilemma was never solved. What happened, instead, was a figment of evolutionary imagination. In the late 1960s, Kimura developed the idea of ‘neutral’ evolution. He reasoned that, if most of the DNA in a cell were non-functional, it would be free to mutate over time. Thus, there would be no cost to the organism to maintain the non-functional portions.
Ohno is credited with inventing the term ‘junk DNA’ about four years later (early 1970’s). The idea of junk DNA is extremely important to evolutionary mathematics. What would happen if it turned out that there was no such thing? What would happen if, instead of being 97% junk, the genome were 97% functional? Even before the completion of the Human Genome Project, though the junk DNA idea was clearly wrong, there seemed to be stubborn refusal to reject junk DNA theory.
Modern technology destroys the concept of ‘junk DNA’.
There are extensive reasons why many today believe that the majority of DNA in the cell is in fact functional. For example, functions have been found for many retrotransposons, 24 of which were once thought to be ‘pieces of viruses’ that had inserted themselves into our genomes over the course of millions of years. Also, functions have been found for much of the vast stretches of non-protein-coding DNA that sits between genes. It turns out that most of the genome is active.
The 2004 (to current) ‘ENCODE’ project was a multi-university, multimillion dollar, multi-year study designed to determine how much of the human genome was transcribed (turned into RNA, a measure of function). ‘Intended as a follow-up to the Human Genome Project, the ENCODE project aims to identify all functional elements in the human genome. The project involves a worldwide consortium of research groups, and data generated from this project can be accessed through public databases. The project is beginning its fourth phase as of February 2017-source: Wikipedia’
From initial phases, which only analysed 1% of the genome, but which included both protein-coding and ‘junk’ DNA regions in the analysis, this project demonstrated that, on average, any given letter of the genome is used in six different RNA transcripts. This does not mean everything is turned into protein, far from it. It also does not mean that everything has a mandatory function or even that the letters are used often. What it means is that almost every letter does something.
Since ‘form follows function’ is a general rule of biology, the fact that these regions are active strongly suggests they have a function. Why else would the cell allow so much transcription? A significant portion of cellular resources are dedicated to making non-protein-coding RNA. Thus, the cell would be greatly benefitted by turning off this waste of energy. Natural selection over millions of years should have killed off the parasitic RNA processing.
But it has not done so because it is actually necessary for cellular function. In fact, the genome can now be seen as an RNA computer (see below). Every year more functions are still being found for the non-protein coding DNA. In fact, the ‘junk’ looks like it is more active than the ‘genes’, turning the old idea that we are protein-based organisms on its head. In the words of an evolutionary biologist, J.S. Mattick:
The failure to recognize the full implications of this—particularly the possibility that the intervening noncoding sequences may be transmitting parallel information in the form of RNA molecules—may well go down as one of the biggest mistakes in the history of molecular biology.
Ultra-complex gene processing
Biology students of the past were always taught the one gene, one enzyme hypothesis. Based on the amazing discoveries over the course of the 20th century, it seemed a straightforward conclusion that a ‘gene’ was a piece of DNA that coded for a specific protein. A ‘gene’ had specific starting and ending points, sections that coded for protein (exons), perhaps a few intervening sequences (introns) that needed to be cut out of the primary RNA transcript before translating the ‘gene’ into a protein, and upstream and downstream regulatory areas where things could bind to the DNA and control the expression of the ‘gene’.
It was easy to walk through a ‘gene’ and see these places. One could even translate the DNA into protein on-the-fly from the sequence of the messenger RNA strand, if one knew the three-letter codes for each amino acid (which is not that hard to do). There is a grave problem with the one gene, one enzyme hypothesis. However, like Darwin’s idea of Pangenesis and the existence of junk DNA, it is also wrong.
A comparison of the programming architecture of the E. coli bacterium to the call graph of the Linux operating system reveals interesting comparisons and stunning differences between the two systems. From the results of the ENCODE project specifically, the world has been given a glimpse inside the most sophisticated computer operating system in the known universe—the human genome. But it is not a protein computer. Actually, the genome is more like an RNA supercomputer that outputs protein.
In a similar way to your computer, which has a hard drive, re-writable memory (RAM), and a screen for output, the genomic computer has DNA for information storage, the RNA for information comparisons and calculations, and protein for output. Efforts have been made to compare genome control processes to human-designed computer systems. The parallels are interesting, but the differences are striking.
For example, when compared to the computer operating system, Linux, the E. coli bacterial genome has fewer high-level regulators, which in turn control fewer middle-level managers, which in turn control many more low-level outputs. It is as if its genome is optimized to do what it does as efficiently as possible. Instead of Linux, a better comparison might be to military computers, which typically have very short programs with minimal instructions. This is true because they are designed to do a limited set of things as efficiently as possible instead of many different things at once (like the simultaneous use of graphics, games, music, and word processing that Linux allows).
Yet, here is another puzzle: it took us many man-hours to design the computers that fly B-52 bombers, yet a single error in any program of any subsystem can cause catastrophic failure of the system it is controlling. The human genome is far more complex and can withstand thousands of errors without crashing. It controls more things simultaneously and is better designed!
When the human genome was completed, researchers were shocked to find only about 23,000 ‘genes’ in the genome. How could this be?The number of proteins which the human body produces is many times more than 23,000. As we already knew this beforehand, the number of human genome ‘genes’ were expected to be far more.
Exons and Introns
The ENCODE project gave us strong hints that the human genome has a huge amount of alternate splicing. Each part of a ‘gene’, we have learned, can be used in multiple different proteins. Somehow, the body knows how to create different combinations of what were thought to be distinct protein-coding genes and splice them together to create several hundred thousand unique proteins.
- Not only that, but different cell types can create different proteins from that complicated process.
- Not only that, but different proteins are produced at different times and, somehow, the cells know what to produce, when to produce it, and under which conditions.
There is something controlling this process, and it is not necessarily in the protein-coding portion of the genome. Imbedded within each ‘gene’ is a series of small codes. Each is only a few letters long, but there are multiple codes at the beginning and end of each exon and intron. They make up what has been called the spliceosome, that is, the part of the genome that controls the complicated process of exon recombination, or gene splicing. The complexity of the spliceosome, indeed the complexity of the entire genome of all eukaryotes, is yet another Achilles’ heel of evolutionary genetics. The genome is too complicated and the ‘target’ for mutation is much too large for known species to survive for millions of years, 33 let alone for them to have evolved in the first place.
A DNA code for protein only?
You may be wondering why I have put the word ‘gene’ in quote marks throughout this chapter? It is because we no longer have a definition of the word. At least, the definition has changed to mean something entirely new for all organisms more complex than bacteria. Genetics has taken a turn toward complexity and the simple, old idea has become outmoded. From here on out, when the word ‘gene’ appears, take it in the classical sense: a gene is a piece of DNA that codes for protein. The only problem with this definition is that any particular piece of DNA might be used in multiple proteins, depending on context.
Hyper-complexity of a four-dimensional genome
Let’s kick it up a few notches and look at another level of complexity. When we sequenced the human genome, we thought we would then understand how the genome works. This was a naïve error. What we had done was sequence the linear string of nucleotides only. This was only the first dimension of a genome that operates in at least four dimensions. What does this mean? Think about DNA. It is a string, a line, which, by definition, is one-dimensional. When the human genome was completed, the junk DNA theory seemed to be supported by the fact that genes were found scattered throughout the genome and there seemed to be no clustering of related functions. “Aha,” they said, “gene order is random, a product of random changes over time.”
The second dimension
However, this was a bit short-sighted, for they were only looking at the first dimension. We just learned about alternate splicing. Here, one part of the genome affects another part, either directly or through RNA and/ or protein proxies. This is part of the second dimension of the genome. In order to draw these interactions, one would need to write out the genome and draw lots and lots of arrow from one place to another. To do this, you would need many sheets of paper, which have two dimensions (height and width).
The second dimension of the genome is extremely complex and includes amongst many others:
- specificity factors,
- enhancers,
- repressors,
- activators,
- transcription factors,
- histone acetylization signals,
- DNA methylation signals,
- post-transciptional regulation of RNA,
- alternate splicing.
It plays a major role in the tight coordination and regulation of the vast network of events that occur both in the nucleus and throughout the cell. In this dimension, the order is not significantly important, for gene regulators have to float around in order to find their targets anyway. Having the target immediately next to the regulator is not necessary.
The third dimension
It is at the next level that things get very interesting. The third dimension of the genome is the 3-D structure of the DNA in the nucleus. At this level, genes are not randomly distributed in the nucleus, but are ordered and clustered according to need. Genes that are used together in series may not be found next to each other on the chromosomes, but when the chromosomes fold, they are often found next to each other in 3-D space, and are also often clustered near a nuclear pore or close to a center of transcription.
The first several hundred letters of the human chromosome is a good example of the first dimension of the genome. This is just a string of As, Ts, Gs, and Cs. Note, however, each letter is in its exact required position:
CTAACCCTAACCCTAACCCTAACCCTAACC
The first dimension of the genome is a string of As, Ts, Gs, and Cs. Thus, something is holding them in place. Since the DNA is equivalent to a huge bundle of string, the parts of that bundle that are buried are difficult to access while other parts are more exposed on the outside or in internal pockets.
Part of the code imbedded within the first dimension affects the 3-D folding of the DNA, which, in turn, affects gene expression patterns. This third dimension is extremely important.
The Fourth dimension
The fourth dimension of the genome involves changes to the first, second, and third dimensions over time. The chromosomes are in a particular shape in the nucleus, but that shape changes during development because different cell types need different complements of genes and other genetic instructions. The shape can change in the short term as cells respond to stimuli and unwrap portions of DNA in order to get at buried genes, only to re-wrap that section when the gene is no longer needed.
There are also changes in the ‘junk’ DNA. For example, a great deal of retrotransposon (a.k.a. jumping genes or mobile elements) activity happens as the brain develops, with pieces of DNA from several different classes (L1, Alu, and SVA) copying themselves and jumping around in the genome of individual brain cells. This helps the various brain cells to differentiate.
Also, liver cells tend to have many chromosomal duplications. The final genome of the various cells in the body is not necessarily the one the fertilized egg cell had when it started dividing, and the shape of the genome varies from cell to cell and through time. These examples are not accidental, but a carefully controlled symphony of genomic changes over four dimensions. I hope you are impressed, for the genome was designed by a master Architect.
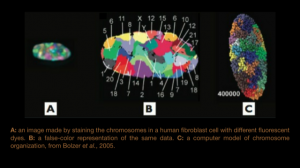
- A: an image made by staining the chromosomes in a human fibroblast cell with different fluorescent dyes.
- B: a false-color representation of the same data.
- C: a computer model of chromosome organization, from Bolzer et al., 2005.
This is all extremely complicated, but it underscores the next Achilles’ heel of evolutionary genetics. Darwinism needs life to be simple. Natural selection needs the ability to take the little tweaks caused by mutation and select the best from a herd or group of animals. Once a species is in existence, perhaps natural selection could work in limited ways, but can this process explain how the species came into existence in the first place?
Hardly, for a simple process of error accumulation and selection could not create a complex, interleaved, four-dimensional system with an amazing amount of data compression and flexibility. And, once that system is in place, it will be seriously threatened by future random changes through mutation. That is the situation we are in today. It is fine to imagine small changes to an already-existing, complex system.
To use those small changes as an explanation for the origin of that system itself, however, is tantamount to saying there was no intelligence involved in the production of the latest computer operating systems. Yet the genome far surpasses in complexity and efficiency any operating system in the world today.
Codon Degeneracy
To date, human-designed computers operate in base-2, because transistors only give us two letters with which to work (0 and 1, off and on), but the resulting math is pretty simple and we have been able to build sophisticated computer chips that operate on this principle. The genome, on the other hand, is base-4. Instead of just 0 and 1, there are four letters in the genome (A, C, G, and T). This is not necessary, for the same information could be spelled out in a system using any number of letters. Why base-4?
Strangely, it takes three letters to code for a single amino acid in a protein. In the genome, these three-letter codons are strung together in groups (exons), with each group producing a functional part of one or more proteins. There are 20 amino acids used in human proteins, but there are 64 possible codons (three positions with four possible letters allows 4 x 4 x 4 = 64 codons). This means that some amino acids, like alanine, are assigned to multiple codons (GCA, GCC, GCG, and GCT). Others, like tryptophan have but one (TGG).
You will notice that the four codons for alanine all start with the letters ‘GC’. This means that any mutation that changes the final letter will produce the same amino acid even though the codon is different. This is referred to as codon degeneracy and it adds a bit of robustness to the genome, for at least some mutations in protein-coding regions have little net effect. I said “little” because the transfer RNAs for the various codons are not found in the cell at the same frequency. There have been cases where a single letter change, even though the amino acid was not changed, created a bad mutation. As the protein was being translated, a pause occurred while the rare tRNA was found. This caused the protein to fold incorrectly, resulting in a malformed enzyme.
More recent data indicates that protein translation in general may be affected by codon usage, at least in bacteria, where the rate of translation depends on the exact codons used. Bacterial cells avoid gene promoters inside their genes. When they occur, translation is often slowed temporarily because the bacterial ribosome tends to stick to promoter sequences.
Thus, having alternate codons available allowed the Designer to intelligently engineer genes with fewer internal contradictions with other gene features. Besides all this, there is a very good reason for codon degeneracy and a base-4 genome. Not only is this the most optimal way to code for 20 amino acids, but it also allows for multiple overlapping codes.
Multiple overlapping codes and data compression.
There is a considerable amount of data compression in the genomes of all higher organisms. Any given section of each of these genomes might be doing several things simultaneously. A single letter of DNA might be part of an exon that is in turn used in twenty different proteins. At the same time, that letter might be part of the splicing code that tells the cell when to produce each of those proteins. That letter also might be part of the histone code that the cell uses to know where to wrap the DNA around certain protective proteins called histones. That letter also might affect the 3-D structure of the DNA. It might be part of the pervasive epigenetic code (Chapter 1), and it might be part of a three-letter codon that is translated into a specific amino acid.
Because the genome is base-4, and because of codon degeneracy, the Designer was able to select from alternate codons when faced with fulfilling multiple simultaneous requirements. Degeneracy allowed Him to overlap multiple genomic commands without having to compromise the design requirements of the proteins. Multiple, overlapping DNA and RNA codes defy naturalistic explanation and make it impossible for natural selection to operate as an agent of long-term evolutionary change.
Multilayered system of regulation
Selection runs into a ‘wall of insurmountable difficulty’ when faced with mutations that affect more than one trait simultaneously. Polyfunctionality (also called pleitropy) means that a given mutation can affect completely unrelated traits (say, color vision, the ability to tolerate garlic, and mitochondrial efficiency, although this would be an extreme example). This is still yet another Achilles’ heel of evolutionary genetics.
How could a simple process of trial and error, always seeking the simplest answer to an environmental problem, create an interleaved and multilayered system of regulation? In fact, this system is one of the wonders of the universe. Without this level of multitasking, the genome would have to be much larger and it might not be possible for DNA-based, multicellular organisms to exist at all without it.
________________________________________________________
Evidence for Genesis in our genes
Some thoughts on relating our newly acquired genetic knowledge back to Genesis:
“Modern genetics has revealed an amazing world of complexity to us, but this is not all it can do, for genetics also gives us the ability to test theories of history. With the many different ancient creation myths in existence, would it surprise you to learn that one in particular is an excellent match for what we have learned through genetics? Would it surprise you to learn about the abundant testimony to Creation, the Flood, and the Tower of Babel in our genes? Because Genesis claims to be a history book, and because it claims an all-encompassing history of humanity, it makes certain and specific predictions about human genetics.
These predictions deal with the creation of two original people (Adam and Eve), a population bottleneck that occurred about 1,600 years later in the time of Noah when the world population was reduced to eight souls, and the predicted single dispersal of humanity from a central point in the Middle East a few hundred years later. We must use caution when approaching this subject, for human science has had a pretty bad track record of getting things right throughout the centuries. There is much we could be wrong about today. However, upon careful and critical analysis of the available data, one can easily see the biblical narrative in our genes.
While people like Francis Collins, the head of the Human Genome Project and a proclaimed evangelical Christian, claim there is no evidence of Adam and Eve, there is little to no evidence that most of these people have thought about what the biblical predictions actually are. The Bible states that all of humanity came from a single couple. This is a profound statement about genetics, for it severely limits the amount of human genetic diversity we should find today. But let’s not get stuck thinking like evolutionists and only consider models that involve a progression back to infinity. Everyone after Adam and Eve has to connect back to them through the normal process of sexual recombination and reproduction, but this is not necessarily true of the founding couple. God could have engineered multiple cell lines into Adam’s testes. Likewise, he could have frontloaded Eve’s ovaries with multiple human genomes, all very different from one another. The early population could have had a remarkable range of diversity.
Alternatively, there may have been but one original genome and Eve was a near-clone (without the Y chromosome), perhaps even a haploid clone, of Adam. Thus, biblical models of human genetic history have many options. Which one should we choose? For theological reasons, I favour Eve being a clone of Adam. Thus, she fell under the curse God applied to Adam and his descendants, and she is related to Christ, the kinsman redeemer. Also, since she was formed from a rib taken from Adam’s side, one might assume the cells, muscles, nerves, blood vessels, and DNA (!) of Adam were used to form her.
This is not mandated by the Bible, and I am open to alternate possibilities, but I lean in that direction. It may come as a surprise to hear it, especially if you have heard or read Francis Collins, but the amount of genetic diversity we observe today could fit neatly into Adam and Eve. In fact, the bulk of it could fit into Adam alone! There are about 10,000,000 places that carry common variations in the human genome. The average person carries somewhere around two or three million of these. That is, there are about three million places where sister chromosomes (chromosomes come in pairs; you have two copies of chromosome 1, for example) have different readings in corresponding locations.
It is not a stretch to think that Adam carried nearly all of these common variants in his genome. Why do people only carry a subset of the total? This is probably due to recursion in their family trees. That is, they have inherited identical copies of different sections of the genome from the same distant ancestor through different lines. Add to this the processes of population growth, contraction, subdivision and mixing over biblical time and we do not need millions of people living in the ancient past to explain current human genetic diversity.
Additionally, most common genomic variants come in two versions. When we find more versions than this, they can be explained by an early mutation (e.g. the most common version of blood type O is clearly a mutation [a broken copy] of the gene that codes for the blood type A allele, 46 and it is found worldwide) or a mutation that occurred post-Babel (e.g. the sickle cell gene common to certain parts of Africa, or the blue-eyed allele common in Northern Europe, or the many variations found in certain immune system genes, which are designed to change rapidly). Thus, since most variation could fit into a single person, it may not be necessary to call for strange models of early biblical genetics.
In the Out of Africa model, evolutionists say that humanity went through a near-extinction bottleneck before our population expanded and eventually left Africa. Why is a bottleneck part of their model? Because they are trying to explain the lack of diversity among people spread across the world. This diversity is much less than they at first assumed, based on ideas of a large population living in Africa for a million years or so. The bottleneck is an ad hoc addition to evolutionary theory, but low diversity is part of the creation model from the start.
Interestingly, and as an aside, animals are different. Humans are exceptionally uniform when compared to most all animal species. Chimpanzees are five to six times more diverse. The common house mouse carries huge variations in their population, including multiple chromosomal rearrangements. Examples of this nature abound in the animal world. Some of this is due to degradation of animal genomes over time. Some of this is also due to the different starting point of animals compared to people. The Bible does not say that God created two of every kind of animal. In fact, one might assume He created a functioning worldwide ecology that included high diversity (as also evidenced from the fossil record) within each kind. It is true that only two of most kinds of animal were on Noah’s Ark, but they were selected from a potentially much larger genetic pool than were people.
Since most of the genomic data available today is human (for obvious reasons), we are far from being able to build a model of genetic history for most animal species, but these are interesting points to consider. Besides the number of common variations found in the human genome, there are an untold number of rare variations as well. These tend to occur only in isolated populations and are an indication of mutations that have occurred since Adam, indeed, since Babel. These include blue eyes in the European population, sickle cell anemia in the African population, and many super rare variations that only occur in a single tribe, family or individual. We share a huge amount of genetic variation. This is an indication that we came from a small population in the recent past.
What we don’t share uniformly (population-level variation) is an indication that our genomes are in rapid decline, with a rapid rate of mutation occurring in all people groups worldwide. More on that below. When one considers the female-specific and male-specific sections of DNA, we can see even more confirmation of Adam and Eve. According to a lot of theory and a lot of experimental evidence, mitochondria (those little subcellular power plants that convert sugar to energy) are only passed on through the female line. Since mitochondria also have their own little genome of approximately 16,569 letters, and since this little genome is also subject to mutation over time, we can use it to build a family tree of worldwide female ancestry.
This led to the announcement of an African Mitochondrial Eve in the evolutionary literature back in 1987. If there were any other females alive millions of years ago, only one managed to pass her genome to all people living today. The date of Mitochondrial Eve is assumed to be in the hundreds of thousands of years before present, but only if one assumes a certain slow mutation rate and common ancestry with chimps.
Using real-world mutation rates gives an age for Eve about 6,000 years ago.
A more recent study demonstrated that mutations in the mitochondrial control region (these account for approximately 2/ 3 of all mitochondrial mutations) occur at a rate of one every other generation. Since Eve’s mitochondrial sequence has been reconstructed and published, in the evolutionary literature, and since most mitochondrial lines are less than 30 mutations removed from the ‘Eve consensus sequence’, and since the most divergent are only 100 mutations removed, the diversity of mitochondrial DNA within the modern human population can easily fit into a 6,000-year/ 200-generation time frame.
We know the sequence of more than 99% of the mitochondrial genome of the original human female. There is no evidence for any other. Why do people believe Eve was just one of many females living in a large population a very long time ago? Because this is part of a model of evolutionary history. It is not grounded in reality, but it gives them a convenient excuse for discounting the biblical prediction that there should be but one female lineage in the world. Like the mitochondrial genome, the male Y chromosome gives us a way to build a family tree of all the men in the world. Y Chromosome Adam supposedly existed a very long time ago as well, but at a different time than mitochondrial Eve.
However, and as above, these conclusions are based on models that assume a lot about human history, population size and mutation rates. With the publication of the revised chimp Y chromosome, and the discovery that it is only 70% identical to the human Y chromosome (even this number is generous since half of the chimp Y is missing), evolutionists are forced to conclude that the Y chromosome has mutated extremely rapidly in human history. Yet, the current human Y chromosomes found in the world are all very similar to one another. The only way to maintain similarity under a high mutation load is to have a very recent common ancestor. Viva Adam! Another measure we can use to test the veracity of Adam and Eve is in linkage data.
During sexual reproduction, the cells in both parents go through meiosis, where the chromosomes they inherited from their parents are mixed together. Thus, when parents pass on their genes, they are actually passing on scrambled versions of the grandparents’ chromosomes. This scrambling, called crossing over causes DNA to be inherited in large blocks. There are sections of the genome that have not crossed over in all of human history (indicating a youthful genome). When two variants are inherited together (because they are close together on the same DNA strand), they are said to be linked.
Linkage has been studied in detail and we have learned some very interesting things from it. First, between two and four common blocks explain most of the blocks in all human populations. In other words, there are only a few original chromosomes and pieces of those chromosomes are still intact. There are only ten thousand or so blocks in the genome, which is easily explained if the human population is only about 200 generations old and there are 1–2 crossing over events per long chromosome arm per generation.
Besides the molecular clock assumption behind most evolutionary studies, the majority also assume that recombination is the same through time and across all geography. This is not necessarily so, however, as we know that crossing over is affected by genetic factors (the PRDM9 gene, specifically) and that variations exist within these factors that affect the rate of crossing over in different individuals. 55 This is a challenge to many prior evolutionary studies, including much of what has been claimed about the evidence for our African origins.
Out of Africa or Out of Babel?
We discussed the Out of Africa theory in relation to Mitochondrial Eve above. In this section, let us simply list the corresponding factors between ‘Out of Africa’ and Genesis. According to the most commonly told evolutionary story, we came from a small population that broke up into smaller bands during a single dispersal event that carried humanity across the globe. This dispersion occurred with three main female lineages and one main male lineage in the recent past. Oh, and it went through the Middle East before it got to Europe, Asia, Australia, Oceania, or the Americas. Every single one of those points is predicted by the Genesis accounts of the Flood (Genesis 6–8), Tower of Babel (Genesis 11) and the Table of Nations (Genesis 9–11).
The difference is in the timing (6,000, 4,500, and ~ 4,000 years ago, respectively, vs. tens of thousands of years ago) and the origin (Middle East vs. NE Africa near the Red Sea56). Yet, the conclusions of the Out of Africa model are driven by the evolutionary starting assumptions. 57 In way of summary, they assume that a molecular clock is in operation, causing mutations to accumulate in all populations at the same rate throughout all time. We have already seen that this is not true. They assume all populations have roughly the same demographics (same birth and death rates, average age of marriage, average number of children, etc.).
They also assume there are no differences in the DNA repair machinery between populations, because this would throw off the molecular clock. Thus, when they find more diversity in Africa, they automatically conclude this is the older population and the source for the rest of humanity. However, what if some African tribes have a different genetic history than the rest? There are many Africans who fit into the average world-type mitochondrial sequence pool. There are others who are much more different. Does this mean these are older sequences, or does it mean these people simply picked up more mutations in their mitochondrial lineage for any number of reasons? Interestingly, a recent study claimed that early African tribes remained small and isolated from one another for thousands of years.
This is a recipe for rapid mutation accumulation and genetic drift. World mtDNA Migration According to the Out of Africa proponents has many parallels with the Genesis account. There are some additional correspondences between ‘Out of Africa’ and Genesis. Generally, humans are believed to have existed in Africa for some millions of years as Homo erectus. Then, without explanation, this pre-modern-human population crashed and nearly went extinct.
The ten thousand or fewer survivors managed to hold on, rapidly evolving into modern man. The population rebounded, diversified, and then some of the genetic lineages managed to escape Africa and conquer the world.
The biblical account starts with two people, Adam and Eve. The population then grows to some unknown number and crashes, being reduced to eight souls, with three reproducing couples, at the Flood, 1,600 years after Creation. The population then rebounds, but they continue to defy God as they did before the Flood, so He intervenes and scrambles their languages at a place called Babel, causing them to go separate ways by clans. Thus, several generations after the Flood, they spread out and conquer the world after the Tower of Babel incident.
There is a nice general concordance between the Out of Africa scenario and Genesis after the evolutionary story is modified to fit the data. Specifically, Out of Africa has had to deal with the lack of diversity among people worldwide (hence, the population crash), and the single dispersal of humanity that is evident in our genes. Stylized Out of Africa scenario compared to a stylized biblical account. Evolutionists believe man existed as Homo erectus for a million years or so in Africa. Then, for some unexplained reason, the species crashed and nearly went extinct. Modern man somehow managed to evolve in this genetically catastrophic event (catastrophic because inbreeding is very, very bad). Then, some of the new genetic lineages that were evolving (through mutation) left Africa to spread out and cover the globe. These lineages happened to all be closely related and nobody knows why the others stayed in Africa.
In the biblical model, the population starts with Adam and Eve, grows to an unknown number, is reduced to eight people 1600 years later, then rebounds. The entire population then spreads out across the globe.
The surprising Neanderthal
What do we do, then, with claims for non-human caveman ancestors? New discoveries in archaeology and genetics have caused evolutionary views on Neanderthals, for example, to shift radically in the past decade.
Neanderthals are now believed to have painted in caves, made musical instruments, had the controlled use of fire, buried their dead in a ceremonial manner with the head pointed toward the rising sun, and hunted the landscape for odd minerals in order to grind them up and use them for makeup (cosmetics). The details of this list are still debated among various evolutionists, but to even bring up one of these ideas, let alone several at the same time, would have been tantamount to evolutionary heresy just a few years ago. Thanks to rapid advances in technology, we now have the ability to pull DNA from some of the best preserved Neanderthal bones.
The field of ancient DNA genetics is problematic, for DNA is a fragile molecule that breaks down rapidly upon the death of an individual. Also, some of the damages are similar to those that occur over the course of time in living individuals. Thus, it is sometimes difficult to tell post mortem DNA decay apart from mutations that occurred in the ancestry of that individual. Another problem is contamination. Since the DNA in the ancient sample is necessarily degraded, any contamination from modern DNA will overwhelm the evidence for ancient DNA.
Researchers are well aware of these problems and have gone to great lengths to overcome them, including treating any new find as a crime scene, with clean room techniques designed to reduce contamination from the people handling the bones. 60 The letters indicate the leading 330 amino acids of FOXP2 protein of human, chimpanzee, gorilla, orangutan, rhesus monkey and mouse. The amino acid sequences show two poly-glutamine stretches (indicated in red) and the two specific mutations (bottom row) which set the human sequence apart from the rest of the presented mammals (the N on position 304 and the S on position 326). The terminal 386 amino acids of FOXP2 are identical in all species and are not shown here. Sequences are as reported in Enard et al.* Enard W. et al., Molecular evolution of FOXP2, a gene involved in speech and language, 418: 869–872, 2002.
What have we learned? Would you be surprised if genetics has thrown evolutionists another curveball? After careful consideration of the issues listed above, modern studies on Neanderthal genetics have come to some surprising conclusions. Early work discovered that they had the same gene (FoxP2) that gives modern humans the ability to speak. Then, they discovered that some Neanderthals carried similar versions of skin pigment genes that cause light skin and red hair, then green eyes and freckles, when they occur in people of European descent. Everything was pointing to them being much more human than once thought, but then, based on mitochondrial DNA retrieved from Neanderthal specimens, the conclusion was made that they probably did not interbreed with the ancestors of modern man because Neanderthal mitochondrial sequences are not found in people living today.
These conclusions were short-lived, however, for the full-length Neanderthal genome (well, 60% of it) was published a short time later. 64 If the sequences are accurate, Neanderthal is not what anyone expected. There is now evidence that they interbred with the direct ancestors of modern people, meaning, according to the biological species concept, that we are the same species. It appears that people living outside Africa carry 3–4% Neanderthal DNA. Interestingly, Neanderthal remains have not been found in Africa, but traces of their genome still persist in the places they did live (and beyond).
The new evolutionary explanation is a modified ‘Out of Africa’ scenario, where there was limited interbreeding between modern humans and Neanderthals as they left Africa and displaced Neanderthals on their way to world domination. But this is a departure from the status quo of only a short time ago. Do you recall how confident the evolutionists were when hitting biblical creationists over the head with the African Origins hypothesis? Much of what they believed is now not supported by their own scientific data. There is an alternate scenario that also fits the data. Instead of two large populations that barely mixed, if the Neanderthal population was much smaller than the main wave of migration, there could have been complete mixing with the same result.
If the modern non-African population is 3–4% Neanderthal, then perhaps their population was but 3–4% the size, or somewhere in between with a different degree of mixing. If they were people, especially if they were a post-Flood population, mixing would be natural, for people do what people do, after all. More recent data showed that Neanderthals across their range were more similar to one another than are individuals within any modern people group.
From Spain to the middle of Russia, Neanderthals look more like one extended family—a human family—that lived on the earth in Europe and Asia after the Flood and were overwhelmed by a later migration of people.
Human vs. chimp
For several decades, we have been hearing, “Humans and chimpanzees are 99% identical.” This is not true. The figure was based on some early experimental evidence that compared sections of known genes to each other, so at least some of our DNA is very similar. But our genomes are less than 2% protein-coding, and many genes are not comparable between the two species. Humans have several hundred protein-coding genes (all tightly integrated into the spliceosome) that are absent in chimps.
There are entire gene families found in humans that are not in chimps. This throws a monkey wrench into evolutionary models, for there have only been a few hundred thousand generations since we were supposedly the same species. How could these brand-new genes arise and be integrated into our complex genomes in such a short amount of time? Time is not the deciding factor for evolution. Evolution is measured in generations, and there have not been that many since the assumed common ancestor.”